.

As part of getting their feet wet in bioinformatics, I have my students search for information about various bioinformatics topics using the tools available through the
NCBI portal. One of the things I decided to have them find is the sequence of amino acids in human insulin, which in its active form, has two chains joined by disulphide linkages . A reasonable place to start is to search for protein amino acid sequences, since insulin is a protein.
But when my students did this they found a
record that starts out:
LOCUS AAH05255 110 aa linear PRI 23-JUN-2006
DEFINITION Insulin [Homo sapiens].
ACCESSION AAH05255
VERSION AAH05255.1 GI:13528924
DBSOURCE accession
BC005255.1KEYWORDS MGC.
SOURCE Homo sapiens (human)
ORGANISM
Homo sapiensEukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini;
Catarrhini; Hominidae; Homo.
and ends with the amino acid sequence:
1 malwmrllpl lallalwgpd paaafvnqhl cgshlvealy lvcgergffy tpktrreaed
61 lqvgqvelgg gpgagslqpl alegslqkrg iveqcctsic slyqlenycn
Each of the letters is an abbreviation for one of the 20 standard amino acids.
For instance m = methionine; a alanine and so forth.
Now some of my students knew from other sources that human insulin actually has 51 amino acids. So, they asked,what's going on? How come the record we pulled up has 110 amino acid residues, not 51? My short answer to them was that insulin is produced in an inactive form, proinsulin, that has to be processed post translation. This is basically correct but as we will see we can get a much deeper understanding of what's going on by carefully exploring the data base records. There are other ways to do this I am sure, but rather than blindly searching lets take a careful look at some of the protein data base records avaialble through NCBI, beginning with the original record my students were finding with the 110 amino acid long polypeptide.
I don't pretend to be bioinformatics expert, but let's go exploring.
Near the top of the record is the line:
DBSOURCE accession BC005255.1
Opening the database source link
, BC005255.1, gives you the information about the source of the protein sequence and you can see that it is predicted from a cDNA sequence made from a mature mRNA. Toward the end of this record is both the cDNA and the amino acid sequence for the protein.
But let's see what else we can glean from the original record. The reference provided near the start of the record is:
AUTHORS Strausberg,R.L., Feingold,E.A et al
CONSRTM Mammalian Gene Collection Program Team
TITLE Generation and initial analysis of more than 15,000 full-length
human and mouse cDNA sequences
JOURNAL Proc. Natl. Acad. Sci. U.S.A. 99 (26), 16899-16903 (2002)
PUBMED
12477932Pulling up the reference, via its pubmed number 1247792, using the above link takes to the abstract of the paper and pdf files for free download-always a nice thing to have. But the first sentence in the abstract gives a clue as to what's going on:
"The National Institutes of Health Mammalian Gene Collection (MGC) Program is a multiinstitutional effort to identify and sequence a cDNA clone containing a complete ORF for each human and mouse gene."
So what do we have here? An open reading frame is the actual amino acid coding region of a gene. That's also where the cDNA comes in because cDNA basically is a DNA sequence for the gene with all the introns stripped out producing using mature mRNA and reverse transcriptase. Basically we have the predicted amino acid sequence for proinsulin from the mRNA.
The record provides us with yet more information.
After a list of references to different regions of the protein, which we will return to later, is a set of annotations after the heading Features.
One of these annotations for the 29th to the 109 amino acid residue reads:
Region 29..109
/region_name="IlGF"
/note="Insulin / insulin-like growth factor / relaxin
family; insulin family of proteins; groups a number of
active peptides which are evolutionary related including
insulin, relaxin, insulin-like growth factors I and II,
mammalian Leydig cell-specific insulin-like peptide (gene
INSL3), and early placenta insulin-like peptide (ELIP)
(gene INSL4), insect prothoracicotropic hormone
(bombyxin), locust insulin-related peptide (LIRP),
molluscan insulin-related peptides 1 to 5 (MIP), and C;
cd00101"
/db_xref="CDD:
28985The little note in this case is telling you that region starting from amino acid 29 through 109 is evolutionarily related to a number of different proteins such as insulin, a protein called relaxin, several growth factors, and interestingly, several insect hormones and even a molluscan peptide.
So we have learned a lot about insulin without even finding the actual 51 amino acid sequence!
But wait there's more!
Just before the amino acid sequence for the predicted amino acid sequence from the open reading frame is this series of lines:
/db_xref="CDD:
28985"
CDS 1..110
/gene="INS"
/coded_by="BC005255.1:60..392"
/db_xref="GeneID:
3630"
/db_xref="HGNC:
6081"
/db_xref="MIM:
176730"
CDS stands for CoDing Sequence and clicking on the CDS link give the sequence of nucleotides including start and stop codons along with the amino acid sequence.
Here is a sample record that shows what's in these sorts or records.
CDD stands for Conserved Domain Database and opening the CDD
link can tell one a lot about what's going on. In this case our protein has two conserved domains and is part of a large family of proteins called IlGF. The little summary tells us that:
"Typically, the active forms of these peptide hormones are composed of two chains (A and B) linked by two disulfide bonds; the arrangement of four cysteines is conserved in the "A" chain: Cys1 is linked by a disulfide bond to Cys3, Cys2 and Cys4 are linked by interchain disulfide bonds to cysteines in the "B" chain. This alignment contains both chains plus the intervening linker region, arranged as found in the propeptide form. Propeptides are cleaved to yield two separate chains linked covalently by the two disulfide bonds."
There still yet is more!
The section of our original record has a link to the INS gene, /db_xref="GeneID:
3630".
Opening this link gives access to further information about the INS gene, first showing this screen:
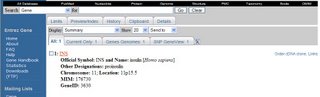
- Official Symbol: INS and Name: insulin [Homo sapiens]
- Other Designations: proinsulin
- Chromosome: 11; Location: 11p15.5
- MIM: 176730
- GeneID: 3630
Note the little tidbit that the INS gene is on chromosome 11 in the upper or P arm.
Open the GeneID link, and look for the various links on the right hand side of that data screen. Open the one labeled "links" and select proteins from the choices. When you do that, you get a whole series of records including one(you may have to scroll down) that says
P01308. This particular record is a Swiss Prot record, and it looked like a fairly complete record related to the proinsulin protein.

Opening
P01308 yields another data record with a wealth of information about the INS gene and the regions of the protein. At the top of this record is a link that says Features. Clicking on that link provides a series of annotations that starts out like the image to the left.
As part of the Feature various regions are annotated. If you have the
P01308 record open and scroll down you will find a region
that says:
Region 1..24
/gene="INS"
/region_name="Signal"
/experiment="experimental evidence, no additional details
recorded".
Click on the Region link, select sequence at the top shows the signalling sequence as:
1 malwmrllpl lallalwgpd paaa
Since the initial product of tranlation includes the signalling sequence, what I have trmed proinsulin is better termed preproinsulin.
Scrolling just a bit farther down in the
P01308 record, brings you to:
Region 25..54
/gene="INS"
/region_name="Processed active peptide"
/experiment="experimental evidence, no additional details
recorded"
/note="Insulin B chain. /FTId=PRO_0000015819."
This is telling you that residues 25 through 54 are the insulin B chain. This chain is 30 amino acids long. Opening this Region link and clicking on Sequence at the top of this record shows you the residues as:
1 fvnqhlcgsh lvealylvcg ergffytpkt
Going back to the
P01308 link and doing a bit more scrolling brings you to
Region 90..110
/gene="INS"
/region_name="Processed active peptide"
/experiment="experimental evidence, no additional details
recorded"
which tells you where the insulin A chain is, namely amino acids 90..110 of our original product from the INS gene.
That region has the amino acid sequence:
1 giveqcctsi cslyqlenyc n.
If you look at the Features, there is a lot more to warm the hearts of your favorite protein chemist about the details of proinsulin's structure!
So getting back to our original protein product from the INS gene we have that:
Residues 1 through 24 is a signalling sequence, residues 25..54 are the B chain; residues 90..110 are the A chain and the remaining part of the protein is a linking region.
Let's see if what we have makes sense with the original amino acid sequence (in red) from way back when:
1 malwmrllpllallalwgpdpaaafvnqhlcgshlvealylvcgergffytpktrreaed
1 malwmrllpllallalwgpdpaaafvnqhlcgshlvealylvcgergffytpkt_____
61 lqvgqvelgggpgagslqplalegslqkrgiveqcctsicslyqlenycn
_______________________giveqcctsicslyqlenycnI did this manually with blue, the signalling region, black the B chain and green the A chain. We still have this linking region to look at:rreaedlqvgqvelgggpgagslqplalegslqkr which comprises amino acids 55-90.
In the record scrolling up to the references there is the following two entries:
REMARK PROTEIN SEQUENCE OF 25-54 AND 90-110.
REFERENCE 10 (residues 1 to 110)
AUTHORS Oyer,P.E., Cho,S., Peterson,J.D. and Steiner,D.F.
TITLE Studies on human proinsulin. Isolation and amino acid sequence of
the human pancreatic C-peptide
JOURNAL J. Biol. Chem. 246 (5), 1375-1386 (1971)
PUBMED 5101771
REMARK PROTEIN SEQUENCE OF 57-87.
REFERENCE 11 (residues 1 to 110)
AUTHORS Ko,A.S., Smyth,D.G., Marktussen,J. and Sundby,F.
TITLE The amino acid sequence of the C-peptide of human proinsulin
JOURNAL Eur. J. Biochem. 20 (2), 190-199 (1971)
PUBMED 5560404
These entries refer to a C-peptide. Scrolling down in the record reveals a series of links to regions and the one for C-peptide give the following sequence of amino acids for the C-peptide:
eaedlqvgqvelgggpgagslqplalegsl
Thus we have mapped out the proinsulin protein as consisting of these regions as before with the C-peptide shown in purple:
1 malwmrllpllallalwgpdpaaafvnqhlcgshlvealylvcgergffytpktrreaed
1 malwmrllpllallalwgpdpaaafvnqhlcgshlvealylvcgergffytpkt**
ead61 lqvgqvelgggpgagslqplalegslqkrgiveqcctsicslyqlenycn
***lqvgqvelgggpgagslqplalegsl***giveqcctsicslyqlenycnSo it looks like the INS gene not only codes for the A and B chains of insulin but also for another polypetide. Looking at the first reference gives the primary structure for this peptide, but what does this peptide do?
At one time it was thought that C-peptide was not biologically active. Indeed some diabetes related sites still talk about it in this way, for instance
this one from Perkinelmer. However, this peptide seems to have a number of effects. For instance a recent paper in Diabetes/Metabolism Research and Review (
abstract) suggests that C-peptide reduces apoptosis of pancreatic islet cells. Another paper in Diabetes/metabolism research and reviews (2003 Sep-Oct;19(5):345-7.,
abstract calls attention to what appear to a number of different biological effects of C-peptide. In some cases the C-peptide clearly works along with insulin. C-peptide seems to increase dilation of arteoles in skeletal muscle in conjunction with insulin as discussed
here. This makes sense since skeletal muscle is a target for insulin. Further there is
some evidence that administration of C peptide improves blood flow in the skin of patients with insulin dependent diabetes.
By the way the C-peptide has
diagnostic value since one C-peptide molecule is released per insulin molecule. Monitering the level of C-peptide can tell doctors about how much insulin the pancreas of a person taking insulin shots is producing.
So what have we got here? The INS gene not only codes for the two insulin chains that are activated later to make insulin, but also codes for another peptide in another violation of the old idea of one gene coding for one polypeptide.
At this point here are some questions that we can investigate:
1. Do the other members of the IlGF also have C-peptides as part of their structure? Doing a quick protein blast of the various protein databases is not particularly illuminating here. So we may have to BLASTn the corresponding nucleotide sequence from the INS gene record.
2. Where did the C-peptide come from evolutionarily? Are there homologous proteins of this type separate from the INS gene? If so ,looking at their function might give insight into other possible functions of the C-peptide. Again the protein databases are not particularly useful here. Maybe the fact that there are no conserved domains, and no apparent C-peptide proteins except for mammals suggest that this protein evolved maybe from a short sequence that got elaborated over time, coevolving with the insulin chains.
3. Many members of the IlGF family are growth factors, what is the tie in between the function of a growth factor and insulin? Looks like a classic case of evolution using material at hand for a new function, but can we infer anything about how that happened?
4. What about other proteins with mutliple chains? Are there analogous peptides to the C-peptide that work along with the multple chained protein? Maybe the one polypeptide product, several protein system from one gene simultaneously is more common than we think.
We are getting into the sorts of areas where more powerful tools such as BLAST and looking at conserved domains can help us with, now that we have dissected the protein product of the INS gene. Now we could have gone to Google and come up with this reference,
http://en.wikipedia.org/wiki/Insulin; but there is something satsfying with getting down with the data.
Other links:
Insulin and Insulin Resistance.
Wilcox G.
Clin Biochem Rev. 2005 May; 26(2): 19-39.http://www.pubmedcentral.gov/articlerender.fcgi?tool=pmcentrez&artid=1204764Specific binding of proinsulin C-peptide to human cell membranes.
Rigler R, Pramanik A, Jonasson P, Kratz G, Jansson OT, Nygren PÅ, Ståhl S, Ekberg K, Johansson BL, Uhlén S, Uhlén M, Jörnvall H, Wahren J.
Proc Natl Acad Sci U S A. 1999 Nov 9; 96(23): 13318-13323.PMCID: 23945
http://www.pubmedcentral.gov/articlerender.fcgi?tool=pmcentrez&artid=23945
Biological activity of C-peptide on the skin microcirculation in patients with insulin-dependent diabetes mellitus.
Forst T, Kunt T, Pohlmann T, Goitom K, Engelbach M, Beyer J, Pfützner A.
J Clin Invest. 1998 May 15; 101(10): 2036-2041.PMCID: 508791
http://www.pubmedcentral.gov/picrender.fcgi?tool=pmcentrez&blobtype=pdf&artid=508791
Technorati Tags:
InsulinC-peptideProteinsGene
 It's fall now, and it looks like Kansas bloggers are spending a lot of time outside and not so much time blogging. So I had to do some serious digging to grub up some posts for this week, but there is still plenty of life and diversity in the Kansas portion of the noosphere and I tried to reflect that diversity in my selections this week.
It's fall now, and it looks like Kansas bloggers are spending a lot of time outside and not so much time blogging. So I had to do some serious digging to grub up some posts for this week, but there is still plenty of life and diversity in the Kansas portion of the noosphere and I tried to reflect that diversity in my selections this week.
















