Indeed there is actually a complex integrin signaling pathway involved in cell development and movement, the details of which can be found on BioCarta. If you are not familiar with this site, it visually shows important metabolic processes and the human genes associated with the proteins involved in these processes. For instance the integrin signaling pathway shown at BioCarta involves an alpha and beta integrin coded for by the genes ITGA1 and ITGB1.
These two integrin subunits appear to be involved in nerve and collagen regeneration.
To try to gain some insight about integrins, I did a blastp using the alpha and beta subunit amino acid sequences for the proteins coded by the ITGA1 and ITGB1 genes. The alpha subunit gave the following conserved domains:

According to the conserved domain summary, the VWA (red)domains appears to be specific to vertebrates and mediate protein interactions between the extracellular matrix and the cell. Significant alignments are found only in chordates with one exception, namely the green sea urchin-not too suprising since Echinoderms and chordates are believed to be closely related.
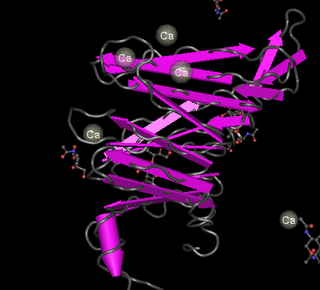
The blue domains are called beta propeller repeats and they are characteristic of integrins. These propellers have a sequence of amino acids that repeats seven times. A beta propeller structure is in this figure made with the Cn3D protein structure viewer. These repeats have regions that apparently bind calcium ions, which is interesting since these ions are often involved in signaling, for instance as part of muscle contraction.
 Since I am always suspicious when a protein seems to have no relatives in bacteria I used NCBI's conserved domain feature to look for related architectures and found 66 references to integrin like proteins in bacteria. The function of these proteins in bacteria is not known but at least some are probably cell membrane proteins. See for instance, Nascimento et al (2004), Journal of Bacteriology, p. 2164-2172, discussing the spirochete Leptospira interrogans. The evidence for this is the presence of seven seven FG-GAP repeats that are highly conserved in the beta propeller repeats of integrins.
Since I am always suspicious when a protein seems to have no relatives in bacteria I used NCBI's conserved domain feature to look for related architectures and found 66 references to integrin like proteins in bacteria. The function of these proteins in bacteria is not known but at least some are probably cell membrane proteins. See for instance, Nascimento et al (2004), Journal of Bacteriology, p. 2164-2172, discussing the spirochete Leptospira interrogans. The evidence for this is the presence of seven seven FG-GAP repeats that are highly conserved in the beta propeller repeats of integrins.As pointed out by this article bacteria often form biofilms consisting of other bacteria and various compounds and perhaps integrins evolved in this context.
For the ITGB1 subunit I found a precursor protein sequence (NP_596867) through NCBI and found there is only one conserved domain called INB which is part of the extracellular component of the polypeptide. Here are the similar domains:

It's interesting that abnormalities in genes for some of the proteins containing some of these domains relate to cellular mechanosensory problems. This makes sense because of the influence the beta subunit has on actin.
The tie in with bacterial proteins gets a bit murky. Most of the related proteins appear to be related to transcription, but one in E. coli may be related to the evolution of it's flagellar mechanism, according to CP Ren et al (J Bacteriol. 2005 Feb;187(4):1430-40)
While there is a lot not known about integrins, it appears that the integrins may not be so unique after all, and that at least in the alpha subunits, we evidence that the genes coding for these proteins can be traced back to the prokaryotes. This is perhaps a similar story to the Homeobox region of the Hox genes involved in animal development being homologous to certain bacterial genes. But that is yet another story, again related to what makes an animal an animal. See the Carroll reference cited below.
References and other links:
Jun Qin, Olga Vinogradova, Edward F. Plow (2004) Integrin Bidirectional Signalling: A Molecular View.
PLoS Biology http://biology.plosjournals.org. 2(6):0726-0729
A good discussion of how integrins send signals into and out of animal cells and the structure of integrins.
E Ruoslahti (1991) Integrins. J Clin Invest. 87(1): 1–5
(http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=294975&tools=bot)
Clear diagram of the types of interactions between cells and their environment mediated by integrins.
INTEGRIN, ALPHA-1; ITGA1; (OMIM 192968)
http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=192968
INTEGRIN, BETA-1; (OMIM 135630) ITGB1(http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=135630)
Carroll, Sean(2005) Endless Forms Most Beautiful, Norton,NY. xi + 350p
I am currently in the middle of this book dealing with the connection between evolution and development or "evo devo". A big theme of this book is that evolution often involves using the same tools in new ways.
Technorati Tags:
Animals
Biology
Proteins
Bioinformatics
No comments:
Post a Comment